|
This section describes how to select the parameter file and to adjust the
most sensitive parameters for a particular experiment. For a detailed
description of all parameters see the
Parameters page. After you have adjusted the parameters and the result seems
to be good, you should save the parameter set to use in the future with similar
experiments. The set you used before can also be retrieved from a saved results
(meshes) file (make sure in this case that you select
.mat filter when loading the file). If saved in a
separate file (.set), the parameters can be edited with any external text
editor.
1. Algorithm
The typical choice is algorithm
4, sometimes 1, and more rarely 2, they are selected by setting
algorithm=1, algorithm=2,
etc. Try algorithm 4 first, unless it is clear a different one should be used.
The algorithm 3 has practically no benefits over the other algorithms, and
therefore has not been tested in the latest versions of the program. The choice
of the algorithm will mostly depends on the type of the image and of the cells.
See Cell outlining algorithms page for more
details.
- For filamentous rod-shaped bacteria algorithm 4 is the only option.
- For extremely small (area < 20 px2) or irregularly shaped
(not rod-shaped) cell algorithm 1 is the only option, you should also consider
runnig it without generating the meshes (getmesh=0).
- For rod-shaped bacteria any algorithm will work.
- Algorithm 4 is the slowest and the best in working with curved cells.
- Algorithm 2 is faster, but fails on elongated cells and requires a
training set. In the distribution of the suite such set is currently only
available for C. crecsentus (these cells have pointed ends, wide septal
regions, are curved).
- Algorithm 1 is the fastest as it does not use the active contour step at
all. This algorithm has poorer performance in separating cells in clusters and
in tracking them in timelapses.
- If you are using algorithm 1, consider using it with
(getmesh=1) or without
(getmesh=0) meshes. Generation of meshes takes
additional time, but may fail for irregularly-shaped cells. A mesh is
required to produce a coordinate system inside, but frequently only the
outline of the cell is sufficient.
2. Default parameter sets
After you have decided what algorithm to use, load the corresponding
predefined set. There are several general sets alg1.set, alg2.set, alg4.set
tested for C. crescentus under the resolution ~ 0.06 µm/px. Some
other sets were tested for other cell types, such as alg2ecoli.set and
alg4ecoli.set for E. coli cells under the same resolution. For other
cells just select one of the standard sets and modify some of the parameters
if necessary.
3. Image segmentation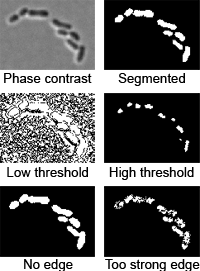
Image segmentation is performed in two steps: thresholding and edge detection:
- Thresholding is detecting the parts of the image above a certain
threshold. The threshold is detected automatically, but the algorithm may
fail if objects other than cells are present or if the background is
non-uniform.
The threshold is multiplied by
thresFactorM
parameter. Default: 1, very high values of the parameter (such that the
threshold exceeds the maximum value for
the image bitdepth) will cause an error. On even images with multiple cells no
adjustment is necessary. Automatic threshold detection may fail on the images
that are uneven, only partially illuminated, have too few cells, or have a lot
of dust particles. You can see that by either keeping the cell-free areas of
the image (the program will typically process and discard multiple regions
with the area of about a few pixels each, in this case increase
thresFactorM), or by not seeing many cells
(decrease thresFactorM).
An alternative to
thresFactorM is
threshminlevel
parameter which defines the fraction of the brightest pixels (between 0 and 1)
that are excluded from the set of values used to calculate the threshold. This
parameter should be included and set in the range 0.05 to 0.1 to eliminate the
effect of bright dust particles, glass chips, or any other objects that may
confuse the automatic threshold detection algorithm.
- Edge/valley detection is detecting boundaries of cells
(detailed description). You can choose
between Laplacian of Gaussian edge detection
(LoG, edgemode=1), valley detection
(edgemode=2), none
(edgemode=0), or both
(edgemode=3). The parameters of the method are:
edgeSigmaL and
logthresh (for LoG),
edgeSigmaV,
valleythresh1 and
valleythresh2
(for valley detection),
crossthresh
(for both).
- Segmentation testing tool. Click the Segmentation button on
the 'Parameter test mode' panel to see segmented image for the current
parameter set before cell detection. Try adjusting the parameters mentioned
above and test the effect. See the
Segmentation testing tool
for more information.
4. Other shape-related parameters
- Area. The program rejects the cells larger and smaller than a
certain thresholds, which are regulated by the
areaMin and
areaMax
parameters and expressed in px2. To adjust these parameters, zoom
on a cell and estimate the area of a cell by counting the pixels along and
perpendicularly to the cell. Typically set areaMax
somewhat larger than the largest cell and areaMin
somewhat smaller than the smallest cell. If on other images some extreme
cells get rejected, click on the largest/smallest detected cell to see its
area and to estimate the area of these extreme cells.
- Smoothing cell. The program uses Fourier smoothing keeping a
predefined number of descriptors, defined by
fsmooth
parameter. Typically for extremely small cells fsmooth
should be ~ 10, for normal-size cells ~20, for spaghetti-like filamentous
cells up to ~200. Values too small don't allow to fit a complex shape of a
cell correctly. Values too large are more tolerable, but may result in a
pixilated outline. Increasing this parameter is one of the ways (the only
way for algorithm 1) to smooth the outline. To smooth the cell using
algorithm 2 you can reduce the number of descriptors by setting
Nkeep lower (from the default of 11 to 7-9),
using algorithm 4 increase the outline rigidity (increase
rigidityRange or
rigidity
parameters).
- scaleFactor.
Use this parameter if you have adjusted well the parameter for a particular
set (keeping scaleFactor=1), and then you only
change the resolution slightly by using variable magnification units on the
microscope. Increase or decrease this parameter proportionally to the
resolution (measured in µm/px).
- Cell diameter (algorithm 4 only). Using algorithm 4 you have the
direct control on two aspects of cell diameter: the absolute values and
flexibility. Note, that you don't set the width exactly, it will still be
adjusted to fit actual cells. The absolute value is regulated by
cellwidth
parameter (mesh generation is optionally regulated by an additional parameter
meshWidth).
Estimate this parameter by counting a typical number of pixels across a cell.
The flexibility is regulated by
wspringconst
parameter. The default wspringconst=0.5 produces a
very small error, which is tolerable for most purposes. However, if the exact
value of the diameter is one of the properties you are trying to measure in your
experiment, you should reduce this parameter to about 0.05 will be small enough
for any purposes. If the image is noisy and the cell diameter variations are
caused by image noise, consider increasing it up to about 2.
- Cell rigidity (algorithm 4 only, for filamentous cells). The
rigidity of filamentous cells is regulated by
rigidityB
(elasticity between nodes, < 10, default: 4) and
rigidityRangeB
(number of affected nodes, < cell length, default: 7) parameters. Low values
make the cell too 'flexible', producing kinks sometimes resulting in errors.
High values smooth kinks too much, so that the program may lose the cell.
5. Aligning the shape
-
Attraction/repulsion. Attraction is pulling the cell
outline into the 'dark' (i.e. cell) areas to fill the cell completely. Repulsion
is retracting the outline from 'light' (background) areas so that it does not
extend from the cell. These effects help to fit better isolated cells, but may
cause the shape to penetrate into the neighbor cell. The two effects are
regulated by
attrCoeff and
repCoeff
parameters. Typical values: from 0 to 1. Usually set them in the range 0-0.2 and
adjust only if necessary. In most cases keep attrCoeff <
repCoeff.
-
Alignment testing tool. The 'alignment' button
on the 'Parameter test mode' panel lets the user to see the process of alignment dynamically.
After activating the mode, click any processing button (such as All frames, This
frame, Range, buttons for manual operations) to see the how alignment happens.
This regime when activated is equivalent to
fitDisplay and
fitDisplay1
parameters present and set to 1. Note, MicrobeTracker still saves the data in
memory and to the disk after each frame (if selected), so be careful to not
erase your data! See the Alignment
testing tool for more information.
 6. Splitting and joining cells 6. Splitting and joining cells
-
Splitting cells is regulated by
splitThreshold
parameter (available in algorithms 2-4). This parameter
defines the minimum septation depth in the profile of integrated phase contrast
intensity (see the image to the right) which triggers cells splitting. If the
condition is met, the program will try to fit new contours to each of the parts
of the cells separated by the septum position. Note, if you split the cell
manually, this condition does not have to be met. Typical values of the
parameter ~ 0.3. The value 1 means that the cell will never be split. If the
cells do not split when they should - reduce
splitThreshold (to 0.12-0.25), if they split too early - increase it (to
0.4-0.5).
-
Joining cells. The program will attempt to join two
cells if the distance between two poles is below
joindist
parameter and the angle between the axes of the cells at those poles is below
joinangle
parameter (in radians). If these conditions are met the program will try to fit
a new shape to the two cells and check the splitting condition for the resulting
cell. Only in this condition fails, the new cell outline will be kept. Note, if
you join the cells manually no check for either of the conditions will be
performed, however the program needs to be able to fit new shape to the both
cells. Typical values are 5 pixels for the distance and ~ 1 radian for the
angle, the values can be easily estimated by looking at a highly zoomed image.
|